Research
Posttranslational protein modifications are powerful tools to reversibly modulate protein function. They allow dynamic control of cellular processes like transcription, DNA repair, cell cycle progression or meiosis without the need of de novo protein synthesis. Besides phosphorylation, methylation or acetylation, the attachment of ubiquitin and SUMO (small ubiquitin related modifier) belongs to most frequently used reversible modifications.
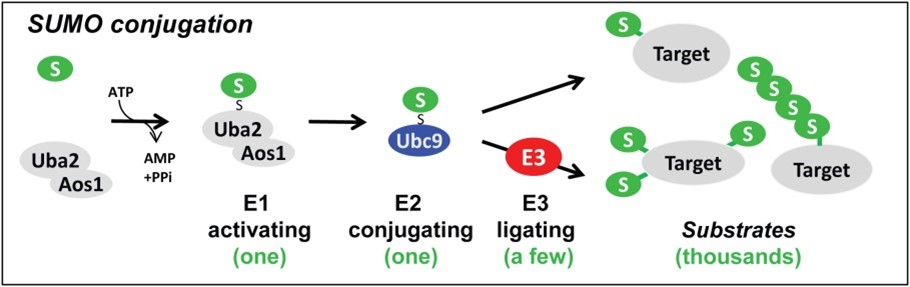
SUMO is a small protein that regulates protein functions like stability, activity, intracellular localization etc. The covalent attachment of SUMO to its substrate, called sumoylation, is essential for viability in most organisms. Sumoylation is executed by the hierarchical action of E1, E2 and E3 enzymes that results in mono- or multi-sumoylation of a target protein or attachment of a SUMO chain (Figure 1). Dysregulation of this system is associated with various diseases ranging from diverse types of cancer to several neuropathological diseases. We aim to understand the molecular mechanisms of how conjugation of SUMO is regulated. In our studies, we attach great importance on biochemical approaches in combination with general cell biology to gain novel mechanistic insights into the powerful complexity of such regulatory SUMO enzymes.
Sumoylation is primarily regulated via E3 ligases and SUMO specific proteases because these enzymes mainly ensure substrate specificity. We characterized an alternative mechanism of regulation on the E2 level: E2 enzyme (Ubc9) sumoylation. This modification is conserved from yeast to mammals but involves structurally different sites of modification, suggesting distinct enzymatic consequences.
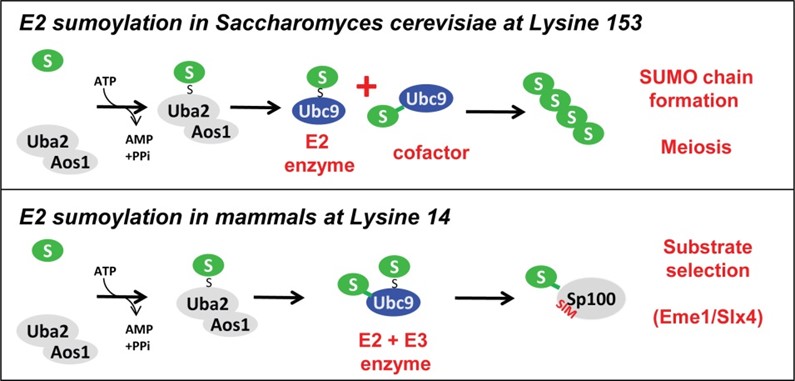
In mammalian cells, we found that N-terminal Ubc9 sumoylation enhances the affinity and modification of selected substrates in dependence of a non-covalent SUMO interaction motif (SIM) (Knipscheer, Klug et al., Mol Cell 2008, Figure 2, lower panel).
By contrast, C-terminal Ubc9 sumoylation in Saccharomyces cerevisiae results in E2 inactivation but turns this inactive Ubc9*SUMO into a cofactor for the unmodified Ubc9. Together, these enzymes cooperate in SUMO chain assembly, which is important for successful synaptonemal complex (SC) formation in yeast meiosis (Klug et al, Mol Cell 2013, upper panel).
We identified and characterized EME1, the regulatory subunit of the structure specific endonuclease EME1-MUS81, as novel substrates for the mammalian sumoylated Ubc9 and are currently investigating how sumoylation regulates their biological functions in dependence on sumoylated Ubc9 and SUMO E3 ligases Nagamalleswari et al, bioRxiv doi: external page https://doi.org/10.1101/2022.04.11.487628. Bakshi et al https://doi.org/10.1101/2022.04.11.487769).
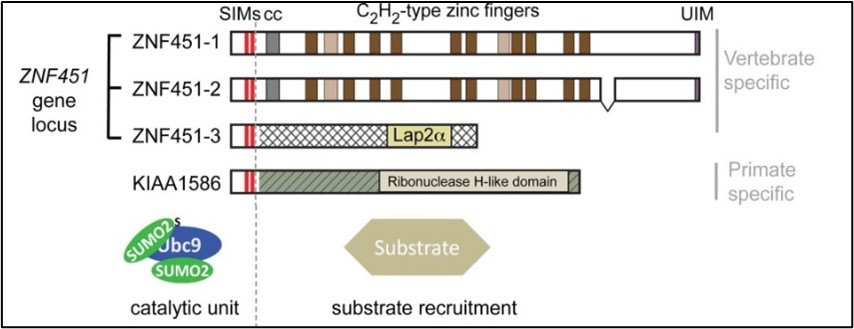
We discovered a novel family of SUMO conjugating enzymes with E3 ligase and E4 elongase (specialized E3s for SUMO chain elongation) functions (Figure 3, upper panel). We have shown that ZNF451 has SUMO E3 ligase activity and efficiently assembles SUMO2/3 chains. Detailed biochemical analysis demonstrated that ZNF451 functions distinct to all known E3 ligases described for SUMO and ubiquitin conjugation: ZNF451 executes catalysis via a tandem-SIM and its interSIM region. One SIM orients the donor-SUMO, while a second SIM binds SUMO on the backside of the E2 enzyme. Intriguingly, this short tandem-SIM region is sufficient to extend a backside-anchored SUMO chain (E4 elongase activity) in contrast to chain initiation, which in addition requires a zinc finger region to recruit the initial acceptor SUMO (E3 ligase activity, Figure 3, lower left panel). Four human proteins share this E4-elongase activity (Figure 3, lower right panel) and are involved in stress-induced global sumo(2,3)ylation after DNA damage induction or proteasome inhibition in vivo (Eisenhardt, Chaugule et al 2015 and Cappadocia et al 2015). We identified novel substrates for ZNF451#1 upon stress stimulation and are currently investigating selected candidates to gain insights into ZNF451#1´s biological function, activation and substrate specificity (Rawat et al, 2021, Urrutia et al 2021, Salas-Lloret D. bioRxiv doi: external page https://doi.org/10.1101/2022.06.22.497173)