Research
Self and non-self DNA in mammalian cells
Extrachromosomal circular DNA (eccDNA) besides mitochondrial DNA exists together with linear chromosomal DNA in every tested eukaryotic cell and can be either be of exogenous origin being introduced during virus or bacterial infections or of endogenous origin like ribosomal circular DNA originating from chromosomes.
EccDNAs also influence the fate of cells. Specifically, in budding yeast extra-chromosomal ribosomal DNA circles accumulates in old mother cells and contribute to their aging. Most remarkably, in mammalian cells the fates as well as cellular effects of such eccDNAs are not well understood.
The Kroschewski lab studies how mammalian tissue culture cells handle intracellular exogenous and endogenous eccDNA. Specifically, we investigate
• their mitotic partitioning
• the underlying mechanisms for eccDNA handling and maintenance in cells to test if there is a relation to cell fate.
Exogenous extra-chromosomal circular DNA
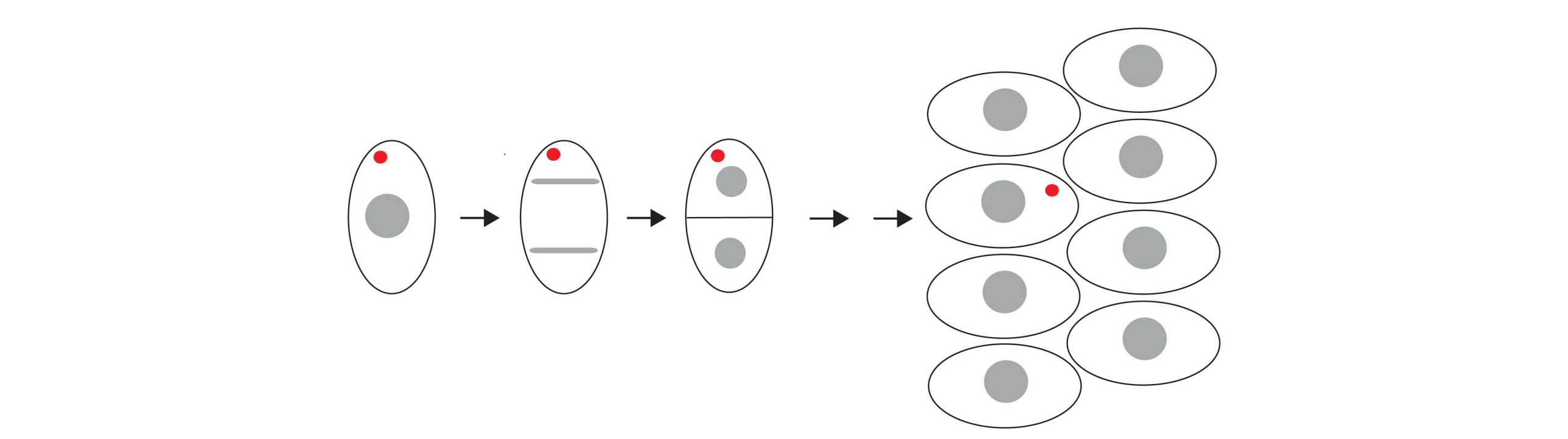
Using plasmid DNA as a model for exogenous DNA (Figure 1) we showed in a recent publication (X. Wang et al, PNAS 2016) that the majority of transfected plasmid DNA is clustered in predominantly in one deposit in the cytoplasm. This deposit is over several divisions asymmetrically partitioned (Figure 2) and is thus maintained in the cell population although in only a few cells. The underlying mechanisms for deposit formation and the biological relevance of this process are unknown.
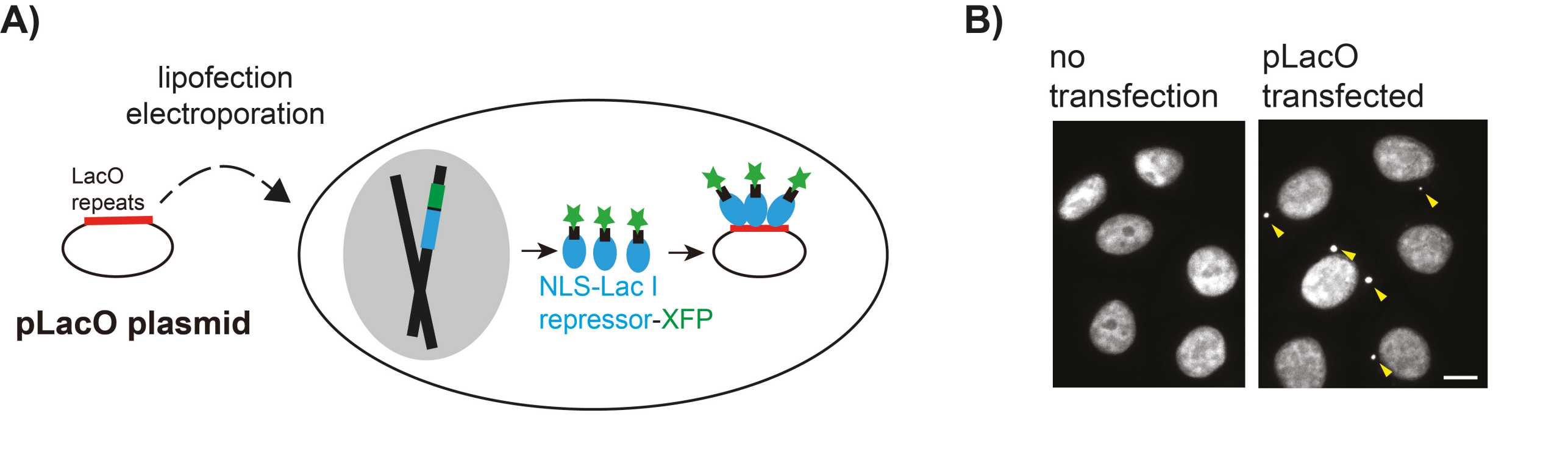
We analyze predominantly HeLa cells stably expressing fluorescently tagged LacI protein and transfected with plasmid DNA containing LacO repeats (pLacO) (Figure 1). Why is that plasmid DNA not completely degraded in cells? What is the mechanism that differentiates transfected plasmid from chromosomal DNA?
Endogenous extra-chromosomal circular DNA
In which cells and under which conditions are endogenous eccDNAs generated? How are they partitioned during divisions? What is their function in cells?
To address these questions we employ the following methods:
Mammalian tissue culture, RNAi- and CRISPR/Cas9-based methods, mass-spectrometry, FISH (Fluorescence in situ hybridization), quantitative light microscopy encompassing confocal fluorescence microscopy or time-lapse microscopy.
Contact
Institute of Biochemistry
Otto-Stern-Weg 3
HPM D11.3
8093
Zurich
Switzerland
