Research
The nuclear pore complex (NPC) is one of the largest protein assemblies in the cell. It is composed of approximately 30 different proteins which are each present in multiple copies, building a complete structure of more than 60 MDa in size (and even up to 120 MDa in multicellular eukaryotes). New developments in cryo-electron microscopy have led to a breakthrough in the determination of the structural arrangement of the NPC, but our understanding of how this large megadalton structure assembles has lagged behind.
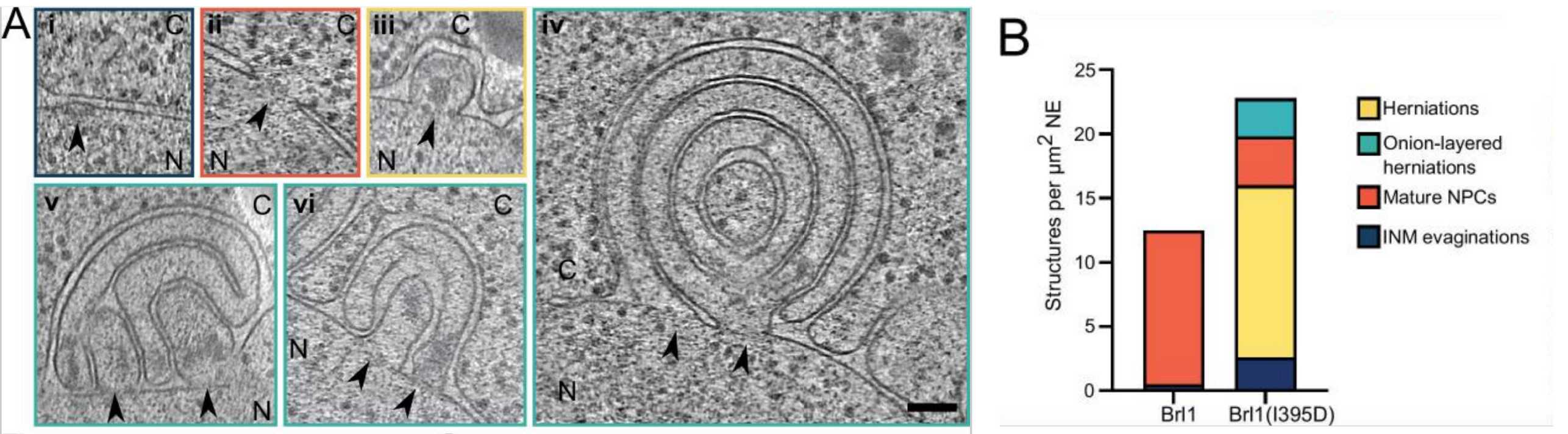
We recently developed a method to systematically characterize the order of events during NPC assembly using advanced mass-spectrometry combined with computational modelling (external page Onishschenko et al., Cell 2020). This work has provided unexpected insight into the timing and molecular sequence of yeast NPC biogenesis. We are now interested in how assembling NPCs are inserted into the double membrane of the nuclear envelope, which requires the inner and outer nuclear membranes to fuse and form a pore. Our results implicate the amphipathic helix of Brl1, a yeast NPC assembly factor, in mediating the fusion step (external page Kralt et al., eLife, 2022), and our future work aims to further dissect the role of Brl1 and associated factors in NPC assembly.
The last step of NPC assembly is binding of the large coiled-coil proteins Mlp1 and Mlp2 which form the nuclear pore basket. Although the nuclear pore basket plays a crucial role in mRNA export, it is present only on a subset of NPCs in yeast cells and its molecular function is not well understood. We have recently developed methods to detect populations of NPCs assembled at different time points and also to visualize individual NPCs in live budding yeast cells (external page Zsok et al., MBoC, 2024). The Dultz group is using this method as well as other approaches to investigate the functional differences between nuclear pore complexes with and without nuclear baskets and during different physiological states.
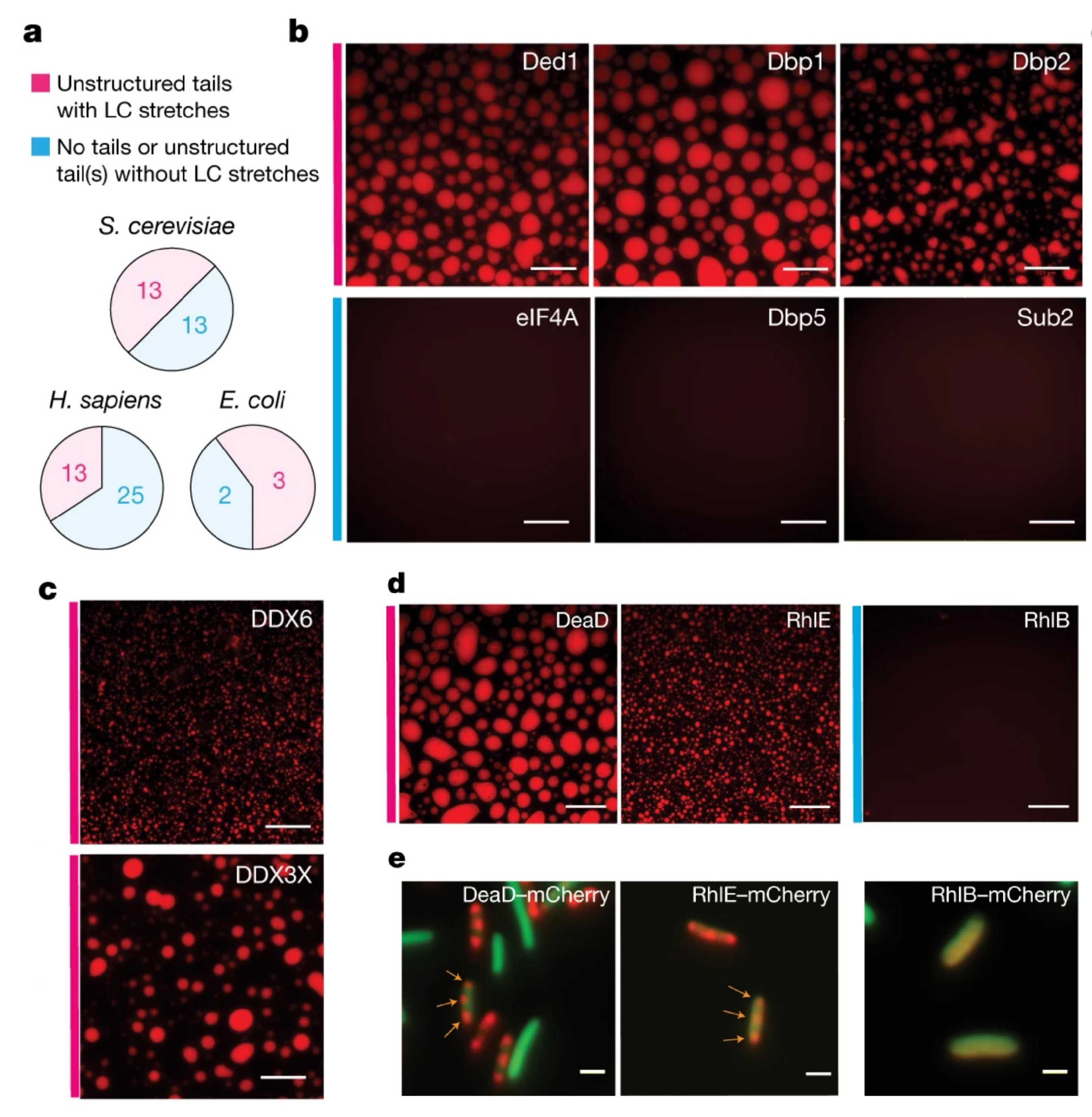
Biomolecular condensates are membraneless organelles that form when proteins and/or nucleic acids undergo liquid-liquid phase separation to form droplet-like compartments distinct from the surrounding dilute phase. Some biomolecular condensates such as nucleoli, splicing speckles and mammalian processing bodies (P bodies) are constitutive, while others such as stress granules and yeast P bodies assemble and disassemble dynamically in response to cellular conditions. Over time, condensates can mature, or ‘harden’, into more solid-like aggregates, a process that has been implicated in several neurodegenerative diseases. Whether this is correlative, causal, protective or pathogenic is unknown, and we are currently trying to understand the function and regulation of biomolecular condensates in health and disease.
One avenue of research aims to determine how biomolecular condensates affect gene expression and cell survival. For example, we have recently shown that the nuclear poly(A) binding protein Nab2 forms nuclear foci following a block in mRNA export, e.g. during glucose stress, and that the material properties of these foci affect the degree of mRNA retention in the nucleus (external page Heinrich et al., Cell Reports, 2024). We are also interested in P bodies, which are cytoplasmic condensates enriched for mRNA degradation factors and mRNA, and in yeast form following various stresses such as glucose starvation and oxidative stress. We have previously shown that the protein Dhh1, a DEAD-Box ATPase and P body component, is a critical regulator of P body dynamics (external page Mugler et al., eLife, 2016; external page Sachdev et al., eLife, 2019; external page Hondele et al., Nature, 2019). We have also shown that disrupting P body formation results in impaired translational repression of many mRNAs following glucose withdrawal in yeast (external page Zedan et al., BioRxiv, 2024). We are currently expanding our in vitro and in vivo model systems to further elucidate the molecular mechanisms underlying the role of Dhh1 in the assembly and disassembly of P bodies, and the effect of their material properties on gene expression.
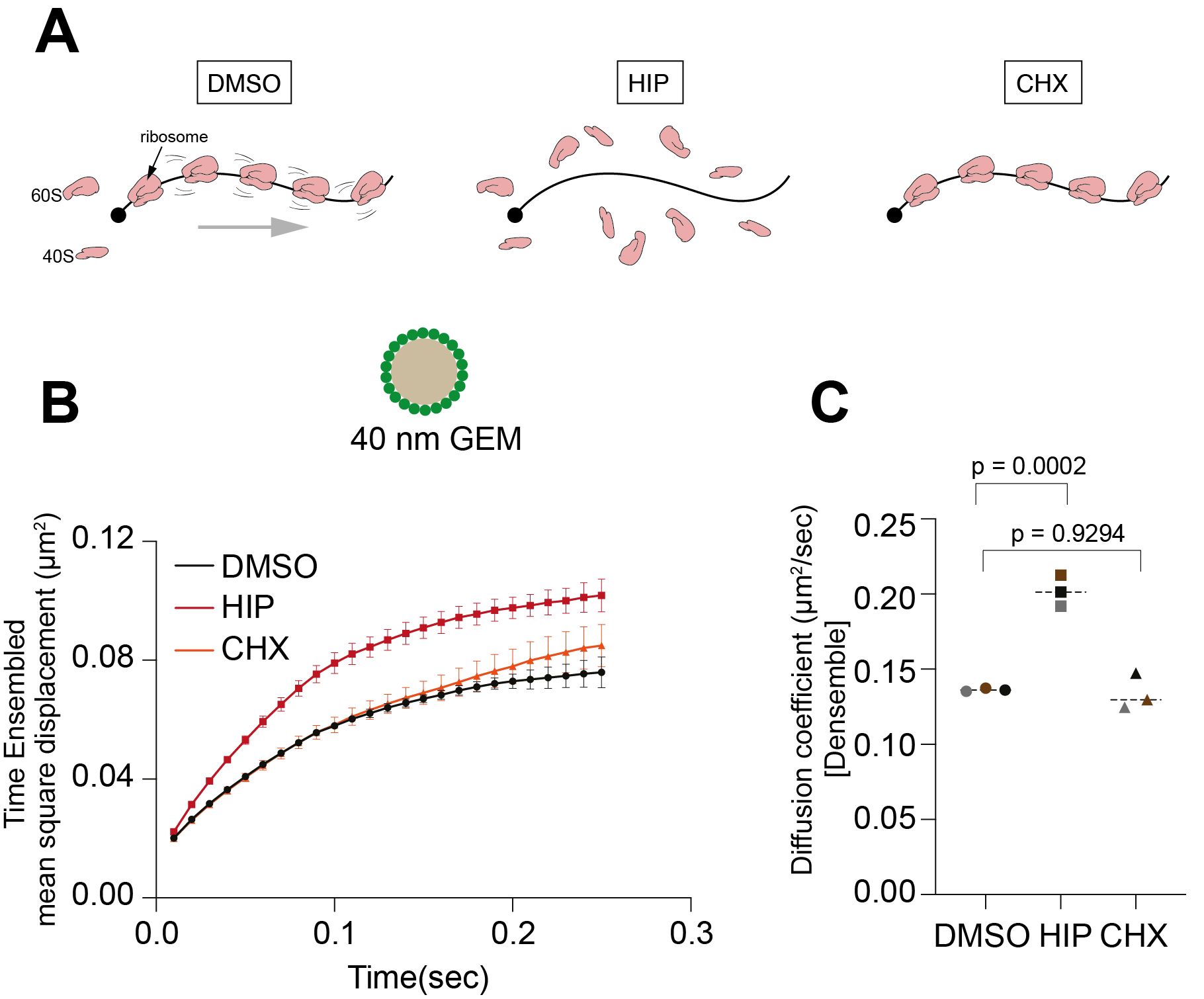
The eukaryotic cell cytoplasm is a highly crowded environment, with organelles, biomolecular condensates, macromolecules and small metabolites all jostling for space. In recent years it has become increasingly apparent that the spatial organisation of the cytoplasm is highly regulated. Changes in growth conditions can induce dramatic changes in the material properties of the cytoplasm, which can fluctuate between a fluid- and a gel-like state. These changes in turn affect the rates of biochemical reactions and can have significant effects on cellular metabolism. We recently showed that the organisation of mRNA into polysomes affects diffusivity and controls the biophysical properties of the cytoplasm (external page Gade et al., BioRxiv, 2024).
Previously, we showed that entering quiescence by glucose starvation in yeast results in a significant restriction in macromolecular movement, brought on by a reduction in cell volume and subsequent increase in macromolecular crowding (external page Joyner et al., eLife, 2016). To further investigate this phenomenon, we performed a genetic screen to identify the molecular players involved in regulating the biophysical properties of the cell during quiescence and chronological ageing. We discovered several genes that, when inactivated, either impair or enhance chronological lifespan. This has opened up multiple avenues of research, and several ongoing projects aim to follow up on the results of this screen. Through these projects, we aim to gain a comprehensive understanding of the genetic, metabolic and biophysical changes that a cell undergoes during stress and ageing.