Research
Regulation of cell growth and division by selective degradation mechanisms
Much of our research is focused on elucidating how cell growth and division are regulated in space and time, in particular by selective degradation of key cellular components. Eukaryotic cells use autophagy and the ubiquitin-proteasome system (UPS) as their major protein degradation pathways. Whereas autophagy is mainly responsible for the degradation of long-lived proteins and entire organelles in the lysosome/vacuole, UPS is used for rapid degradation of proteins when fast adaptation is needed. E3-ubiquitin ligase complexes are important components in the UPS pathway, since they specifically select the relevant ubiquitination substrates. In particular, cullin-ring based E3-ligases (CRLs) emerged as critical regulators of cell division, and many aspects about their composition and substrate-specific adaptors have been elucidated. However, little is known about their regulation, and the identity and function of their key substrates. Autophagy pathways selectively remove protein aggregates and damaged or excess organelles, but little is known about the targets and mechanisms that provide specificity to this process. Available results suggest that ubiquitin not only regulates proteasomal degradation, but may also be critical to ensure specificity in autophagy, and ubiquitin signaling, thus ongoing efforts focus on the molecular connections between autophagy and UPS pathways.
We are thus using biochemistry, structural biology, modern yeast genetics, RNAi-screening, dynamic and quantitative light microscopy and microfluidic technology to tackle the three main topics described below both in yeast and mammalian cells.
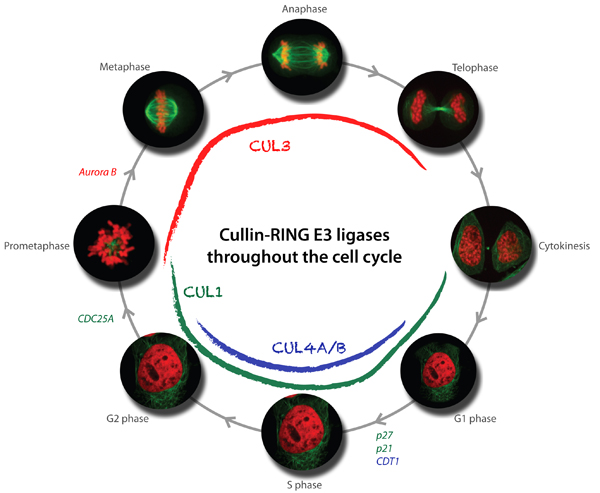
Function and regulation of cullin-based E3-ubiquitin ligases
Available results strongly indicate that cullin-based E3 ligases are critical regulators of cell cycle progression. In particular, our genetic and biochemical studies in yeast, C. elegans and mammalian cells revealed that multiple cullin3-based E3-ligases (CRL3) temporally and spatially control mitosis, and are required for correct chromosome alignment, attachment and segregation, proper midzone and midbody formation and completion of cytokinesis. Moreover, cullin4-based E3-ligases (CRL4) are required for accurate and complete DNA replication, which is of fundamental importance to maintain genome integrity. Using yeast genetics and RNAi-based screening approaches, we have identified several substrate-specific adaptors regulating cell cycle transitions, and ongoing experiments are aimed at the identification of critical substrates. Moreover, we have also identified novel functions of cullin-based E3-ligases outside cell cycle regulation, and characterize these processes at the molecular level.
In addition to the functional analysis, we are interested in the mechanisms involved in the regulation of cullin-based E3-ligases. The assembly and activity of CRLs is regulated by covalent attachment of the UBL-protein Nedd8 (neddylation) of the cullin subunits, and we study the components and regulation of the neddylation machinery and its downstream consequences for cullin function. Through genetic screens in yeast and C. elegans we identified a conserved component termed Dcn1 (Defective for Cullin Neddylation), which is required for cullin neddylation and CRL activity in vivo. Structural and biochemical analysis suggests that Dcn1p functions as a scaffold-like E3 ligase for neddylation. Indeed, the 1.9 Å X-ray crystal structure of yeast Dcn1 revealed an N-terminal ubiquitin binding (UBA) domain and a C-terminal domain of unique architecture, which directly interacts with cullins through a conserved DAD-patch motif. As DCNL proteins appear to function as E3-ligases, we isolate DCNL-interacting proteins to identify novel neddylated proteins.
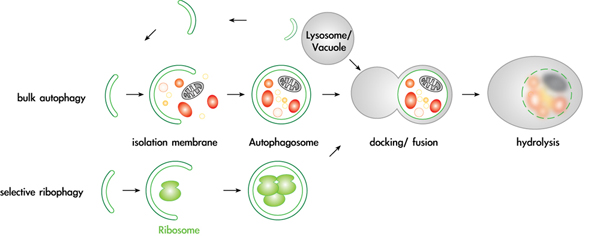
Mechanisms and regulation of selective autophagy pathways
Autophagy is an essential pathway during nutrient starvation, and is also required to selectively eliminate proteins, protein aggregates and damaged or excessive organelles. It has recently become evident that defects in autophagy are causally involved in numerous diseases, including cancer and neurodegenerative disorders such as Alzheimer’s and Parkinson’s disease. However, despite this intriguing medical potential, surprisingly little is known about the fundamental cellular mechanisms, in particular about the physiological targets and mechanisms that provide specificity to this process. We recently found that several proteins and entire organelles such as mature ribosomes are rapidly degraded by selective autophagy upon nutrient starvation. A genetic screen revealed that selective degradation of ribosomes requires catalytic activity of the Ubp3/Bre5 ubiquitin protease. Indeed, ubiquitination of several ribosomal subunits and/or ribosome-associated proteins was specifically enriched in ubp3Δ cells, suggesting that the regulation of ribophagy by ubiquitination may be direct. These results uncovered an intriguing link between ubiquitination and regulated degradation of mature ribosomes by autophagy. We currently investigate the regulation and function of ribophagy in yeast and mammalian cells, and initiated several genetic and biochemical approaches to identify components and regulators of selective autophagy pathways. In particular, we are determining the regulation and physiological substrates of the protein kinase Atg1, and examining putative autophagy receptors.
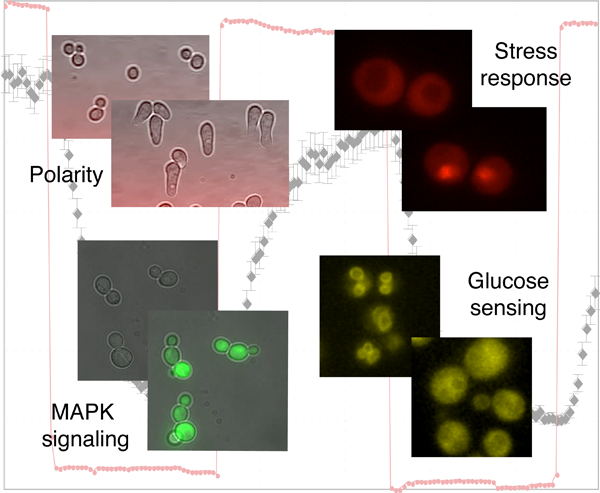
Dynamic and quantitative analysis of signal transduction pathways in single cells
A general characteristic of living organisms is their capability to adapt to different conditions. To accomplish this task, they have engineered complex signaling pathways to sense their intra- and extracellular environment and trigger an appropriate response. Many signals are transmitted by conserved mitogen-activated protein kinase (MAPK) modules, which respond to various stimuli such as stress conditions, growth factors or cytokines binding to G-protein coupled receptors. Due to the wide range of stimulus transmitted through MAPK signaling cascades, alterations in these pathways have been linked to numerous diseases including cancer, diabetes or inflammatory diseases. While genetic and biochemical analysis unraveled the components involved in signal transduction, we combine semi-automated microscopy and microfluidic techniques to obtain quantitative measurements of signaling dynamics at the single cell level. These data uncover novel regulatory mechanisms that ensure the fidelity and robustness of signal transduction and establish the necessary quantitative measurements required to generate testable mathematical models.